Single Cell
The adult mammalian heart is a highly heterogeneous organ. Whole-transcriptome RNA-sequencing of bulk heart tissue reveals considerable insight into disease biology, but is limited in resolution, thus important biological information is obscured.
Single-cell RNA sequencing (scRNA-seq) offers the ability to capture whole transcriptomic profiles of individual cells and cell nuclei, at unprecedented resolution. This permits the identification and study of specific cell types and sub-populations, in a global manner that does not necessitate antibody-epitope detection or the construction of genetic marker tracing models. Read more about this in our recent commentary on the topic, here.
We recently adapted the established Fluidigm microfluidic platform for single nuclear RNA-seq (snRNA-seq). snRNA-seq of mouse and human failing and non-failing adult hearts revealed considerable cellular heterogeneity, and the presence of cardiomyocyte sub-populations with altered expression of cell-cycle related genes. Read more, here.
This technique may be applied to any snap-frozen tissue sample, including banked human tissue.
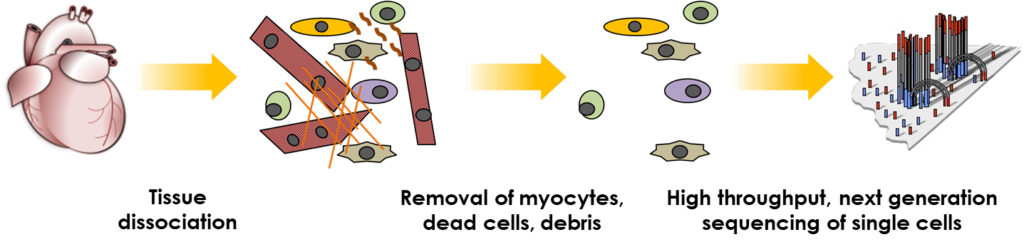
We have also optimised flow cytometry-based procedures (FACS-seq) to replace the requirement for any commercial microfluidic platforms. FACS-seq offers tailored solutions with greater flexibility and reduced cost, and may be applied to both single cell (fresh sample) and single nuclei (fresh or frozen sample) sequencing. However, in common with microfluidic platforms, FACS-seq is not suited for scRNA-seq of live adult cardiomyocytes, due to their large size, poor survival and fragmentation during the sort procedure.
Add Your Comment